Chargaff rule. Chargaff's_rules 2020-01-14
Chargaff's_rules
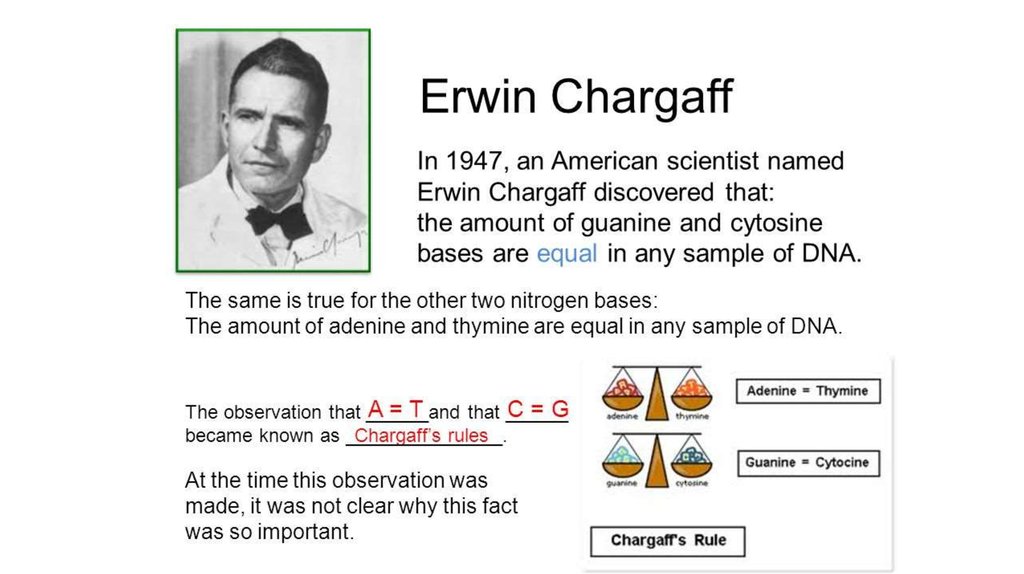
The Z curve database: a graphic representation of genome sequences. Asymptotically increasing compliance of genomes with Chargaff's second parity rules through inversions and inverted transpositions. . In 2006 it was shown that this rule applies to four of the five types of double stranded genomes; specifically it applies to the eukaryotic chromosomes, the bacterial chromosomes, the double stranded viral genomes, and the archeal chromosomes. On the desoxyribonucleic acid content of sea urchin gametes. Insights into the evolution of organellar genomes.
Next
Chargaff's_rules

Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. The composition of the deoxyribonucleic acid of salmon sperm. This seems likely to be the result of Szybalski's and Chargaff's rules. Because the number of purine bases will to a very good approximation equal the number of their complementary pyrimidines within the same strand and because the coding sequences occupy 80-90% of the strand, there appears to be 1 a selective pressure on the third base to minimise the number of purine bases in the strand with the greater coding content and 2 that this pressure is proportional to the mismatch in the length of the coding sequences between the two strands. J Biol Chem, 186 1 :37-50.
Next
Chargaff's_rules

J Biol Chem, 195 1 :155-60. These codons normally differ in the third codon base position. J Cell Physiol Suppl, 38 Suppl. The rule itself has consequences. Szybalski, in the 1960s, showed that in bacteriophage coding sequences A and G exceed C and T. While Sybalski's rule generally holds, exceptions are known to exist.
Next
Chargaff's_rules

Queen's University, Kingston, Ontario Canada. Albrecht-Buehler has suggested that this rule is the consequence of genomes evolving by a process of inversion and transposition. Because of the computational requirements this has not been verified in all genomes for all oligonucleotides. A test of Chargaff's second rule. In most bacterial genomes which are generally 80-90% coding genes are arranged in such a fashion that approximately 50% of the coding sequence lies on either strand.
Next
Chargaff's_rules

It has been verified for triplet oligonucleotides for a large data set. Multivariate statistical analysis of codon use within genomes with unequal quantities of coding sequences on the two strands has shown that codon use in the third position depends on the strand on which the gene is located. The separation and estimation of ribonucleotides in minute quantities. The basis for this rule is still under investigation. J Biol Chem, 192 1 :223-30. Because of the asymmetry in pyrimidine and purine use in coding sequences, the strand with the greater coding content will tend to have the greater number of purine bases Szybalski's rule. The longer the strands are separated the greater the quantity of deamination.
Next
Chargaff's_rules

The has 64 of which 3 function as termination codons: there are only 20 amino acids normally present in proteins. Deviations from Chargaff's second parity rule correlate with direction of transcription. Some recent studies on the composition and structure of nucleic acids. Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Biochem Biophys Res Commun, 340 1 :90-94. The origin of the deviation from Chargaff's rule in the organelles has been suggested to be a consequence of the mechanism of replication.
Next
Chargaff's_rules

Chemical specificity of nucleic acids and mechanism of their enzymatic degradation. Composition of the desoxypentose nucleic acids of four genera of sea-urchin. The biological basis for Szybalski's rule, like Chargaff's, is not yet known. Cold Spring Harbor Symp Quant Biol, 31:123-127. The combined effect of Chargaff's second rule and Sybalski's rule can be seen in bacterial genomes where the coding sequences are not equally distributed. The mismatch between the number of codons and amino acids allows several codons to code for a single amino acid. This process tends to yield one strand that is enriched in G and T with its complement enriched in cytosine C and adenosine A and this process may have given rise to the deviations found in the mitochondria.
Next
Chargaff's_rules

This process does not appear to have acted on the mitochondrial genomes. . . . . . .
Next
Chargaff's_rules

. . . . . . .
Next
Chargaff's_rules
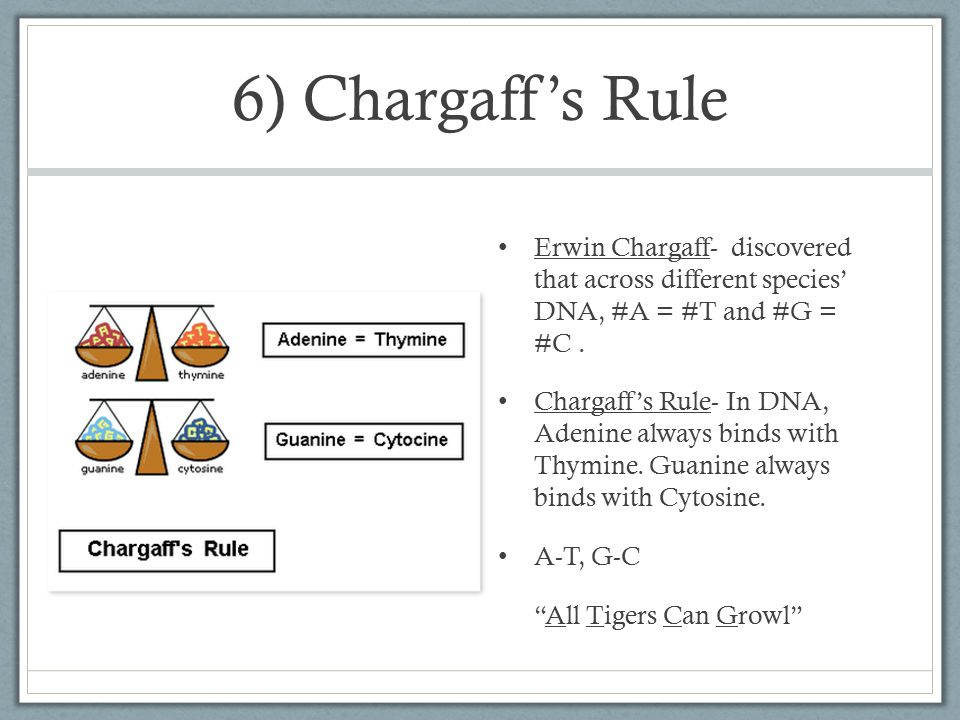
. . . . . . .
Next









